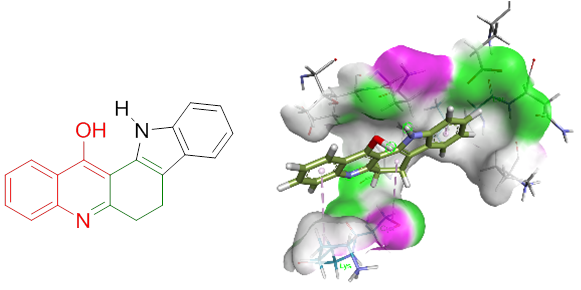
Design, synthesis and molecular docking studies of indolo[2,3-a]acridinol derivatives
Аннотация
A study of the synthesis of an indolo[2,3-a]acridinol derivative using the Claisen ester condensation reaction resulted in the discovery of inexpensive and user-friendly solvents. Structures of the newly synthesized compounds were characterized by FT-IR, 1H NMR, 13C NMR, and HRMS analyses. Docking studies showed a strong affinity of indolo[2,3-a]acridinol towards prostate cancer-related proteins. The binding affinity closer to 10 kcal/mol indicated effective binding. Indolo[2,3-a]acridinol showed strong binding affinities towards protein androgen receptors such as 1GS4, 1T7R, 2AX8, and 3B66 indicating its potential role in protein kinase inhibition. The programs, AutoDock 4 and AutoDock Vina, and Swiss ADME software were applied to dock the target protein with synthesized compounds.
Литература
Suzuki H., Ueda T., Ichikawa T., Ito H. Endocr.-Relat. Cancer 2003, 10, 209-216. https://doi.org/10.1677/erc.0.0100209
Ni M., Chen Y., Lim E., Wimberly H., Bailey S.T., Imai Y., Rimm D.L., Liu X.S., Brown M. Cancer Cell 2011, 20(1), 119-131. https://doi.org/10.1016/j.ccr.2011.05.026
Zelefsky M.J., Morris M.J., Eastham J.A. Cancer of the Prostate, Williams & Wilkins, 2019.
American cancer society, https://www.cancer.org/cancer/prostate-cancer/about/what-is-prostate-cancer.html (date of access 21.07.2022)
Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. CA Cancer J. Clin. 2015, 65(2), 87-108. https://doi.org/10.3322/caac.21262
Lacivita E., Perrone R., Margari L., Leopoldo M. J. Med. Chem. 2017, 60, 9114-9141. https://doi.org/10.1021/acs.jmedchem.7b00965
Ali I. Current. Can. Drug. Targets 2011, 11, 131-134. https://doi.org/10.2174/156800911794328457
Haider R., Ahmad K., Siddiqui N., Ali Z., Akhtar M.J., Fuloria N., Fuloria S., Ravichandran M., Shahar Y.M. J. Bioorg. Chem. 2019, 88, 102962. https://doi.org/10.1016/j.bioorg.2019.102962
Rømer M.U., Nygård S.B., Christensen I.J., Nielsen S.L., Nielsen K.V., Müller S., Smith D.H., Vainer B., Nielsen H.J., Brünner N. Mol. Oncol. 2013, 7(1), 101-11. https://doi.org/10.1016/j.molonc.2012.09.001
Zhao B., Liu P. Nat. Commun. 2020, 11, 908. https://doi.org/10.1038/s41467-020-18046-y
Matiadis D., Sagnou M. Int. J. Mol. Sci. 2020, 21(15), 5507. https://doi.org/10.3390/ijms21155507
Wang W., Ho W.C., Dicker D.T., MacKinnon C., Winkler J.D., Marmorstein R., El-Deiry W.S. Cancer Biotherapy 2005, 4, 893-898. https://doi.org/10.4161/cbt.4.8.2134
Nepolraj A., Pitchai P., Mani P. Org. Chem. Res. 2019, 5, 167-173. https://doi.org/10.22036/ORG.CHEM.2019.151260.1171
Nepolraj A., Pitchai P., Vijayarathinam M. Malaysian J. Chem. 2019, 21, 66-72.
Nepolraj A., Shupeniuk V.I., Sathiyaseelan M., Prakash N. Vietnam J. Chem. 2021, 59, 511-521.
Shupeniuk V.I., Nepolraj A., Tarasa T.N., Sabadakha O.P. Russ. J. Org. Chem. 2021, 57, 582-588. https://doi.org/10.1134/S1070428021040126
Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Adv. Drug. Deliver Rev. 2001, 46, 3-26. https://doi.org/10.1016/s0169-409x(00)00129-0
Daina A., Michielin O., Zoete V. Sci. Rep. 2017, 7, 42717. https://doi.org/10.1038/srep42717
Meng X.Y., Zhang H.X., Mezei M., Cui M. Curr. Comput-Aid. Drug 2011, 7, 146-157. https://doi.org/10.2174/157340911795677602
Forli S., Huey R., Pique M.E., Sanner M.F., Goodsell D.S., Olson A.J. Nature Protocols. 2016, 11, 905-919. https://doi.org/10.1038/nprot.2016.051
Saeed A., Ur-Rehman S., Channar P.A. Drug. Res. 2017, 67, 596-605. https://doi.org/10.1055/s-0043-113832
Saeed A., Mahesar P.A., Channar P.A. Chem. Biodivers. 2017, 14, e1700035. https://doi.org/10.1002/cbdv.201700035
Willard L., Ranjan A., Zhang H. Nucleic. Acids. Res. 2003, 31, 3316-3319. https://doi.org/10.1093/nar/gkg565